Vortex problem with oscillating boundaries#
In this tutorial we will consider a two-dimensional moving vortex problem inspired by the Taylor-Green vortex problem, introduced in [HW07]. The problem is implemented in the file MovingVortex.py, but note that it has not been optimized for parallelization because of its intended use for code verification.
Problem description#
The problem has an analytical solution of the two-dimensional incompressible Navier–Stokes equations in the absence of body forces, \(\mathbf{f} = 0\), namely:
where \(\mathbf x = (x_1, x_2)\) are the Euclidean coordinates, and \(\nu = 0.025\) \(\text{m}^2\)/s. Furthermore, the mesh velocity \(\mathbf w\) is determined by the following displacement field:
where \(\mathbf X= (X_1, X_2)\) are the ALE coordinates, \(A_0=0.08\) is the amplitude, \(T_G=4T\) is the period length of the mesh motion, \(T\) is the length of one cycle, and \(L=1\).
Simulation in OasisMove#
To simulate this flow problem in OasisMove using the default parameters you can run the following command:
$ oasismove NSfracStepMove problem=MovingVortex
and the simulation will start. The simulation should only take a couple of seconds when running with the default parameters, as it is run on a relatively coarse mesh for only 20 time steps.
Results#
When the simulation is finished, there will be a folder named results_moving_vortex, in the current working directory.
The results-folder contains all simulation results per run. Running the moving vortex problem with the default
parameters should produce three solution files located in the Solutions
folder: velocity.xdmf, pressure.xdmf, and the exact velocity solution velocity_exact.xdmf. These solution files
can be visualized in ParaView or any similar visualization software, and in
Fig. 7 we display the displaced grid, velocity and pressure solution at \(T=1\).
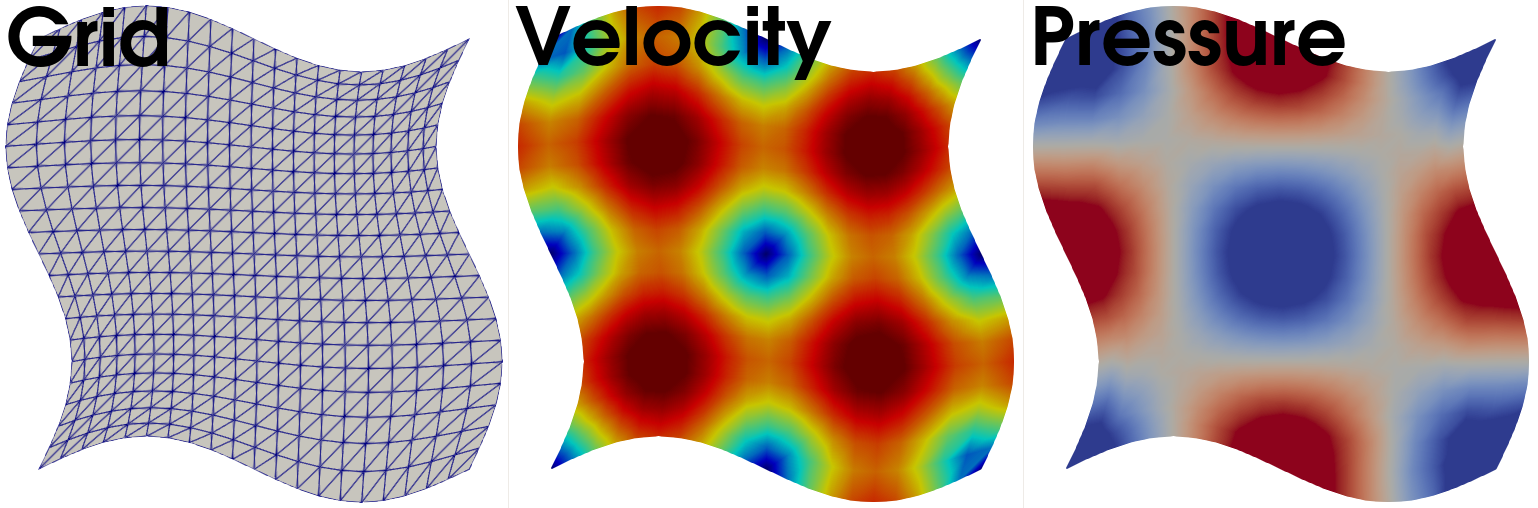
Fig. 7 From left to right: The grid displacement, velocity solution and pressure field at \(T=1\).#
Increasing the spatial and temporal resolution#
The default resolution for the vortex problem is using the resolution parameters \(N_x=20\) and \(N_y=20\) resulting in \(2\times N_x \times N_y = 800\) triangular cells in the mesh. To increase the mesh resolution, we can supply these parameters as command-line arguments to change their value. For instance, to create a mesh consisting of 20000 cells, we can run the following command:
$ oasismove NSfracStepMove problem=MovingVortex Nx=100 Ny=100
Similarly, we can adjust the number of time steps the simulation should perform by adjusting the time step
parameter dt, which by default is \(\Delta t = 5\cdot 10^{-2}\). To simulate 1000 time steps between \(t=0\) and \(t=T=1\),
we can run the following command:
$ oasismove NSfracStepMove problem=MovingVortex T=1 dt=0.001
Important
When adjusting spatial and temporal resolution is important to know relationship between the cell size and the time-step size, which are closely related through the Courant number (\(C\)) given by the Courant-Friedrichs-Lewy (CFL) condition:
where \(u\) is the velocity magnitude, \(\Delta t\) is the time step size, and \(\Delta x\) is the length interval. The general consensus is that \(C_{\max} = 1\).
- HW07
Jan S Hesthaven and Tim Warburton. Nodal discontinuous Galerkin methods: algorithms, analysis, and applications. Springer Science & Business Media, 2007.
